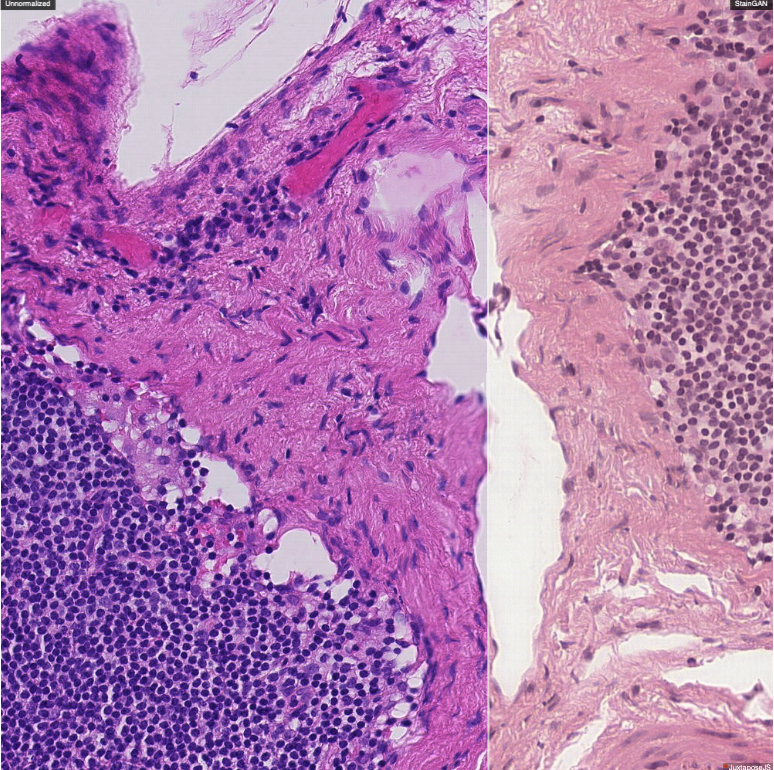
StainGAN
StainGAN implementation based on Cycle-Consistency Concept
For more information visit website.
Structure
- Stain-Transfer Model
- Pre-processing.
- Post-processing.
- Evaluation
Datasets
The evaluation was done using the Camelyon16 challenge ( https://camelyon16.grand-challenge.org/) consisting of 400 whole-slide images collected in two different labs in Radboud University Medical Center (lab 1) and University Medical Center Utrecht (lab 2). Otsu thresholding was used to remove the background, Afterwards, 40, 000 256 × 256 patches were generated on the x40 magnification level, 30, 000 were used for training and 10, 000 used for validation from lab 1 and 10, 000 patches were generated for testing from lab 2.
Patches can be found here: https://campowncloud.in.tum.de/index.php/s/iGgQ9vdHiMZsFJB?path=%2FStainGAN_camelyon16
Any use of the dataset or anypart of the code should be cited
Citation
If you use this code for your research, please cite our papers.
@inproceedings{shaban2019staingan,
author = {Shaban, M Tarek and Baur, Christoph and Navab, Nassir and Albarqouni, Shadi},
booktitle = {2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019)},
organization = {IEEE},
pages = {953--956},
title = {Staingan: Stain style transfer for digital histological images},
year = {2019}
}
Acknowledgments
Code is inspired by pytorch-DCGAN and CycleGAN.