Computational Medical Imaging Research (Albarqouni Lab)
Clinic for Diagnostic and Interventional Radiology
University Hospital Bonn
Faculty of Medicine
University of Bonn
The Albarqouni Lab, based at the
Clinic for Diagnostic and Interventional Radiology at the
University Hospital Bonn, conducts research at the intersection of medical imaging, artificial intelligence, and collective intelligence.
The lab’s mission is to develop the next generation of computational imaging algorithms that improve clinical decision-making while remaining privacy-preserving, robust, and globally accessible.
Inspired by principles of distributed intelligence, the lab views AI systems not as isolated models, but as collaborative ecosystems—where knowledge is learned locally, shared responsibly, and aggregated to benefit patients and healthcare systems worldwide. This perspective directly informs the lab’s work in federated learning, affordable AI, and trustworthy medical imaging.
The Albarqouni Lab is affiliated with the Munich School for Data Science (MUDS), the Medical Imaging Center Bonn (MIB), and the European Laboratory for Learning and Intelligent Systems (ELLIS).
Research Focus Areas
Research at the Albarqouni Lab is structured around three complementary pillars:
-
Computational Medical Imaging
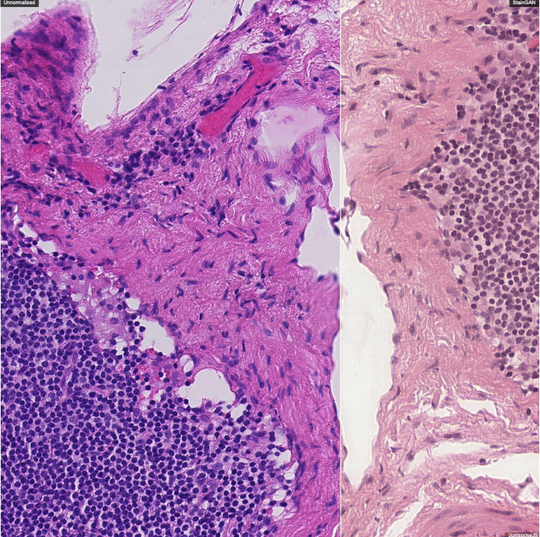
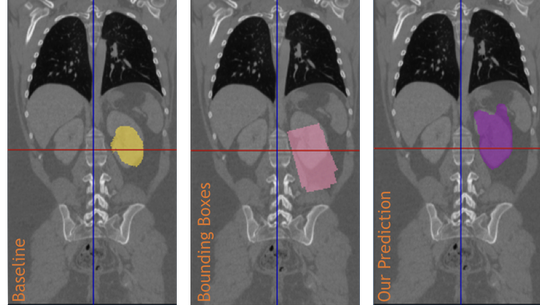
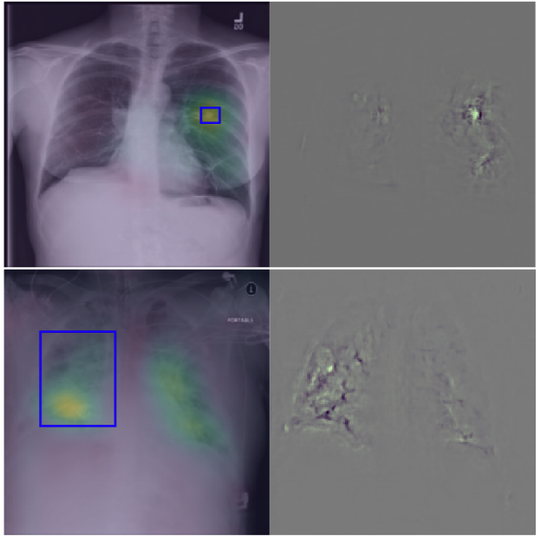
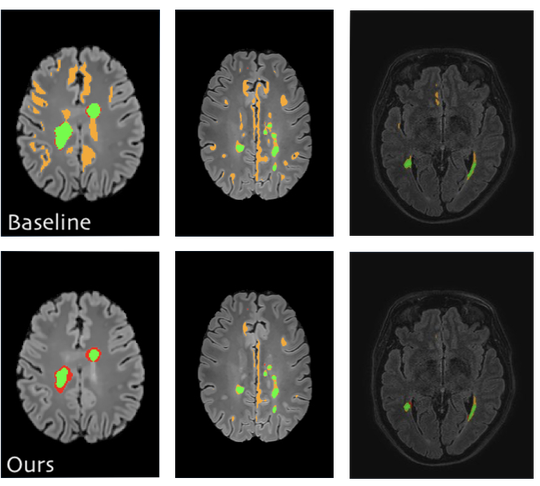
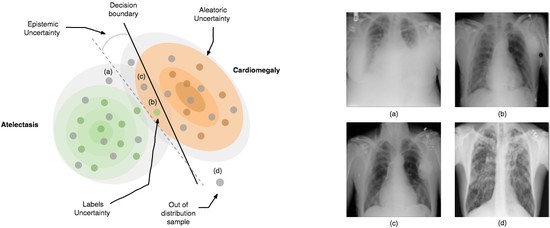
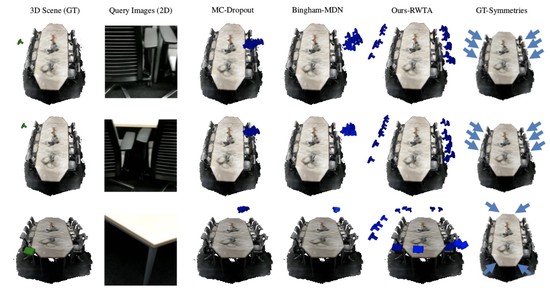
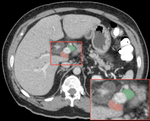
The lab develops automated and data-efficient imaging methods that enhance accuracy, robustness, and reproducibility in clinical workflows. Key challenges addressed include limited and noisy annotations, inter- and intra-observer variability, class imbalance, scanner heterogeneity, and domain shift across institutions. -
Federated Learning in Healthcare
The lab advances decentralized learning frameworks that enable multiple institutions and AI agents to learn collaboratively without sharing raw data. Research topics include robustness to data heterogeneity, explainability, quality control, resilience to adversarial or poisoning attacks, and reliable deployment in real-world clinical environments. -
Affordable AI for Global Health
A core commitment of the lab is to design AI solutions that remain effective under resource constraints, including low-quality data, limited infrastructure, and point-of-care settings. This work aims to ensure that advances in medical AI are inclusive, scalable, and impactful beyond high-resource healthcare systems.