MA Thesis: Deep Learning based model for detection and grading of prostate cancer using mpMRI and MR-Fingerprinting -- Not available
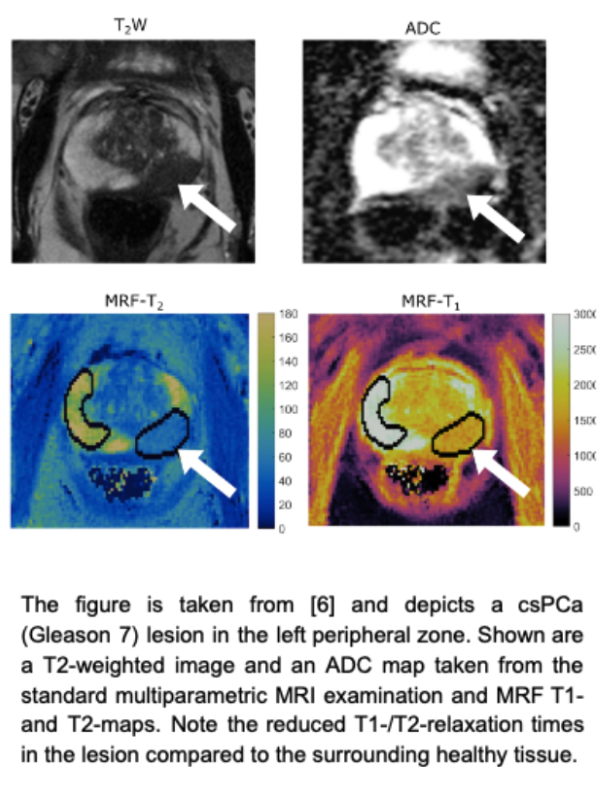
Abstract. Prostate cancer (PCa) is the most common cancer in men and the second leading cause of cancer death in Germany [4,14]. Both digital rectal examination (DRE) along with the prostate-specific antigen (PSA) level in blood samples are typically used in PCa screening. Altogether, about 40% of males in Western industrialized countries have the risk of developing PCa during their lifetime, of whom only about 10% become symptomatic and 3% die [15,16]. To determine the clinical significance of PCa, prostate biopsies are assessed histologically. Prostate lesions with a pathology/histology of Gleason score ≥ 7, and/or volume ≥ 0.5 cc, and/or extraprostatic extension (EPE) are commonly considered clinically significant Prostate Cancer (csPCa). Readers are referred to [17,18] for further details about the acquisition and the interpretation.
Treatment options of active surveillance, surgery, and/or radiotherapy are determined, accordingly. Multiparametric magnetic resonance imaging (mpMRI) has been increasingly utilized for the detection and staging of csPCa. The PI-RADS 2.1 scoring system [1][2] was introduced to standardize the image acquisition and interpretation (scoring) of csPCa. Recently, Alice et al. [5], Lo et al. [6], and Panda et al. [13], among others, have shown that quantitative characterization of prostate lesions can be successfully performed using diffusion MRI and Magnetic Resonance Fingerprinting (MRF) [3]. MRF represents an MRI sequence with a novel data acquisition, post-processing, and visualization approach. A pseudorandomized acquisition pattern with the variation of flip angle, repetition time and echo time within a scan allows the measurement of specific signal patterns, so-called “fingerprints”, which, via matching with a database (“dictionary”), enable the simultaneous generation of co-registered, multiparametric quantitative maps, based on T1-, T2- and T2-relaxation times. In this project, we will investigate developing a data-driven deep learning-based model to automatically detect and stage the csPCa cases using the MRF-based relaxometry, and potentially combined it with the mpMRI examination, including high b-value imaging and apparent diffusion coefficient mapping (ADC).
Objectives:
- Given a database of morphological high-resolution T2w images (in axial and sagittal orientation), diffusion MRI images including a high b-value image of 1500 and ADC mapping as well as MRF sequences and their corresponding pixel-wise delineation of the Prostate lesions, the PI-RADS v2.1 scoring (1-5), and histopathological grading - if available - we aim to develop a deep learning (DL) model to:
- segment the prostate lesions
- automatically detect and stage the prostate lesions in csPCa, and
- eventually, predict the histopathological grading (Gleason score) – if time allows
- Run a comparative analysis between the DL models trained on T2w, high b-value DWI, and ADC, and the one trained on MRF sequences - potentially combined with the high b-value DWI and ADC image.
- Report relevant evaluation metrics such as the Dice Coefficient, and Area Under the Precision-Recall Curve (AUPRC).
Dataset:
- Cohort: Up to date, 171 patients with elevated PSA levels have been included in this ongoing study.
- MRI: Patients received an MRI examination on a 3T MRI scanner (Philips Ingenia) including the following sequences:
- High-resolution T2-weighted sequences in axial and sagittal orientation
- DWI, including b-values of 100, 400, and 800 with the calculation of an ADC map, and a high b-value of 1500
- Magnetic Resonance Fingerprinting (MRF)
- Perfusion imaging
- Pre- and post-contrast T1-weighted axial images of the pelvis
- Data annotation: Axial high-resolution T2-weighted images are labeled by a radiology resident with 4 years of experience to include the following labels: i) Peripheral Zone, ii) Transitional Zone, iii) Lesion
- Histopathological evaluation: Patients that were graded with a PI-RADS score of 3 or higher will receive a systematic/targeted biopsy that allows histopathological correlation with lesions.
Roadmap:
- Familiarize yourself with the current literature on
- Radiomics model for prostate [10] and its repeatability with MRF [12]
- Deep Learning with MRF in Parameter Estimation [7-9], Correlation with Histopathology [11], and lesions catheterization [13]
- Develop the baseline and proposed method
- Run extensive experiments and analysis
- Write up your thesis
Requirements:
- Solid background in Machine/Deep Learning
- Familiar with discriminative deep learning models and SOTA architectures
- Sufficient knowledge of Python programming language and libraries (Scikit-learn)
- Experience with a mainstream deep learning framework such as PyTorch.
- Machine/Deep learning hands-on experience
References:
- Steiger, P. and Thoeny, H.C., 2016. Prostate MRI based on PI-RADS version 2: how we review and report. Cancer Imaging, 16(1), pp.1-9.
- Turkbey, B., Rosenkrantz, A.B., Haider, M.A., Padhani, A.R., Villeirs, G., Macura, K.J., Tempany, C.M., Choyke, P.L., Cornud, F., Margolis, D.J. and Thoeny, H.C., 2019. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. European urology, 76(3), pp.340-351.
- Ma, D., Gulani, V., Seiberlich, N., Liu, K., Sunshine, J.L., Duerk, J.L. and Griswold, M.A., 2013. Magnetic resonance fingerprinting. Nature, 495(7440), pp.187-192.
- Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I, Bray F (2020). Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Available from: https://gco.iarc.fr/today, accessed [01 July 2022]
- Alice, C.Y., Badve, C., Ponsky, L.E., Pahwa, S., Dastmalchian, S., Rogers, M., Jiang, Y., Margevicius, S., Schluchter, M., Tabayoyong, W. and Abouassaly, R., 2017. Development of a combined MR fingerprinting and diffusion examination for prostate cancer. Radiology, 283(3), p.729.
- Lo, W.C., Panda, A., Jiang, Y., Ahad, J., Gulani, V. and Seiberlich, N., 2022. MR fingerprinting of the prostate. Magnetic Resonance Materials in Physics, Biology and Medicine, pp.1-15.
- Hoppe, E., Körzdörfer, G., Würfl, T., Wetzl, J., Lugauer, F., Pfeuffer, J. and Maier, A.K., 2017. Deep Learning for Magnetic Resonance Fingerprinting: A New Approach for Predicting Quantitative Parameter Values from Time Series. GMDS, 243, pp.202-206.
- Girardeau, S., Oksuz, I., Cruz, G., Vasquez, C.P., King, A. and Clough, J., 2019, April. Deep Learning for Magnetic Resonance Fingerprinting. In International Conference on Medical Imaging with Deep Learning–Extended Abstract Track.
- Golbabaee, M., Chen, D., Gómez, P.A., Menzel, M.I. and Davies, M.E., 2019, May. Geometry of deep learning for magnetic resonance fingerprinting. In ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (pp. 7825-7829). IEEE.
- Abdollahi, H., Mofid, B., Shiri, I., Razzaghdoust, A., Saadipoor, A., Mahdavi, A., Galandooz, H.M. and Mahdavi, S.R., 2019. Machine learning-based radiomic models to predict intensity-modulated radiation therapy response, Gleason score and stage in prostate cancer. La radiologia medica, 124(6), pp.555-567.
- Shiradkar, R., Panda, A., Leo, P., Janowczyk, A., Farre, X., Janaki, N., Li, L., Pahwa, S., Mahran, A., Buzzy, C. and Fu, P., 2021. T1 and T2 MR fingerprinting measurements of prostate cancer and prostatitis correlate with deep learning–derived estimates of epithelium, lumen, and stromal composition on corresponding whole mount histopathology. European radiology, 31(3), pp.1336-1346.
- Fujita, S., Hagiwara, A., Yasaka, K., Akai, H., Kunimatsu, A., Kiryu, S., Fukunaga, I., Kato, S., Akashi, T., Kamagata, K. and Wada, A., 2022. Radiomics with 3-dimensional magnetic resonance fingerprinting: influence of dictionary design on repeatability and reproducibility of radiomic features. European Radiology, pp.1-10.
- Panda, A., Obmann, V.C., Lo, W.C., Margevicius, S., Jiang, Y., Schluchter, M., Patel, I.J., Nakamoto, D., Badve, C., Griswold, M.A. and Jaeger, I., 2019. MR fingerprinting and ADC mapping for characterization of lesions in the transition zone of the prostate gland. Radiology, 292(3), p.685.
- Krebs in Deutschland für 2017/2018. 13. Ausgabe. Robert Koch-Institut (Hrsg) und die Gesellschaft der epidemiologischen Krebsregister in Deutschland e.V. (Hrsg). Berlin, 2021
- Bott SR, Birtle AJ, Taylor CJ et al. Prostate cancer management: (1) an update on localised disease. Postgrad Med J 2003; 79: 575-580. doi:10.1136/pmj.79.936.575
- Leitlinienprogramm Onkologie (Deutsche Krebsgesellschaft, Deutsche Krebshilfe, AWMF). Interdisziplinäre Leitlinie der Qualität S3 zur Früherkennung, Diagnose und Therapie der verschiedenen Stadien des Prostatakarzinoms, Langversion 5.1, AWMF Registernummer: 043/022OL. http://www.leitlinienprogramm-onkologie.de/leitlinien/prostatakarzinom/
- Engels, R.R., Israël, B., Padhani, A.R. and Barentsz, J.O., 2020. Multiparametric magnetic resonance imaging for the detection of clinically significant prostate cancer: what urologists need to know. Part 1: acquisition. European urology, 77(4), pp.457-468.
- Israel, B., van der Leest, M., Sedelaar, M., Padhani, A.R., Zamecnik, P. and Barentsz, J.O., 2020. Multiparametric magnetic resonance imaging for the detection of clinically significant prostate cancer: what urologists need to know. Part 2: interpretation. European urology, 77(4), pp.469-480.
Interested, please contact Prof. Dr. Shadi Albarqouni