MA Thesis: Deep Learning based detection model of the temporal and axillary artery in suspected giant cell arteritis in ultrasound images
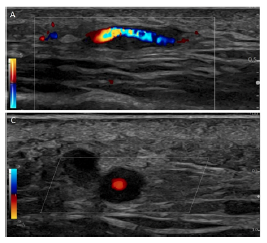
Abstract. Giant cell arteritis (GCA) is a systemic autoimmune disease marked by inflammation of blood vessels (“vasculitis”) that can cause impairment and damage to organs [1]. GCA typically affects large and medium size arteries, such as the aorta and the temporal and axillary arteries [2–4]. It is considered the most common form of systemic vasculitis in adults [5–8], older than 50 years, in Europe [5,9,10] and the USA. Every week, we diagnose one to two new patients with GCA in our department. A diagnosis of GCA must be made as soon as possible through ultrasound verification or falsification as it is a severe rheumatologic and ophthalmologic emergency that can cause permanent vision loss in up to 50% of patients [11,12]. One way to early diagnose GCA is by observing whether hypoechoic wall thickening of the superficial temporal artery is present and measurable in Ultrasound (US) imaging (Fig.1). US devices are widely available and easily accessible because they are noninvasive, patient-friendly, and cost-effective [13]. Moreover, they offer a superior image resolution of less than 0.1mm when used in modern transducers [14]. Although the US can be used to detect GCA [15], expertise in the interpretation of the resulting images is still lacking, and ultrasound specialists are not readily available for the GCA diagnosis.
Aim. To this end, we aim to develop a deep learning-based model which detects the temporal and axillary artery wall thickening in suspected GCA in US images, basically classifying the images whether abnormal or not. Such a model would have an impact on the throughput and precision of the US diagnosis of GCA.
Research Questions:
-
Would unsupervised learning, e.g, anomaly detection models, deliver acceptable performance compared to the supervised models, trained on a few amount of annotated data, which is prone to overfitting?
-
Do the results of the off-the-shelves Interpretability tools; e.g., uncertainty quantification, and visualization methods, e.g., class activation maps (CAMs), correlate with the findings/annotations reported by the US specialists?
Dataset:
- We expect to deliver an amount of 1000 GCA ultrasound images of the temporal artery and 1000 GCA-negative ultrasound images of healthy individuals at a minimum. Starting from an already pre-existing stock of hundreds (> 500 patients) of ultrasound images on GCA and healthy controls and being equipped with three high-end ultrasound machines (GE Logiq S8 and E10) at our site and five to ten ultrasound examinations on patients suspected of having GCA alone each week will help the fulfillment of our goal. In addition, US images would also be retrievable from an international GCA expert group we are part of (the “OMERACT ultrasound subgroup on large vessel vasculitis”). Saving videos instead of just motionless images during future artery ultrasound examinations at our site would generate additional “leverage”, by allowing for later video footage decomposition into single ‘frames per second’ (thereby yielding up to 30 images per second from an ultrasound video), aside from commonly used data augmentation techniques.
Roadmap:
- Familiarize yourself with the current literature [15-20]
- Build the baseline supervised model and develop the anomaly detection model.
- Run the necessary comparisons.
- Equip the models with the Monte-Carlo Dropout [21] for uncertainty estimation.
- Equip the models with the visualization methods, e.g., INNvitstigate [22-23]
- Run extensive experiments and analysis
- Write up your thesis
Requirements:
- Solid background in Machine/Deep Learning
- Familiar with discriminative deep learning models and SOTA architectures
- Sufficient knowledge of Python programming language and libraries (Scikit-learn)
- Experience with a mainstream deep learning framework such as PyTorch.
- Machine/Deep learning hands-on experience
References:
- Weyand CM, Goronzy JJ. Medium- and Large-Vessel Vasculitis. New England Journal of Medicine. 2003;349(2):160-169. doi:10.1056/NEJMra022694
- Aschwanden M, Kesten F, Stern M, et al. Vascular involvement in patients with giant cell arteritis determined by duplex sonography of 2x11 arterial regions. Annals of the rheumatic diseases. 2010;69(7):1356-1359. doi:10.1136/ard.2009.122135
- Lazarewicz K, Watson P. Giant cell arteritis. BMJ. 2019;365:l1964. doi:10.1136/bmj.l1964
- Weyand CM, Goronzy JJ. Giant-Cell Arteritis and Polymyalgia Rheumatica. New England Journal of Medicine. 2014;371(1):50-57. doi:10.1056/NEJMcp1214825
- Watts RA, Robson J. Introduction, epidemiology and classification of vasculitis. Best practice & research Clinical rheumatology. 2018;32(1):3-20. doi:10.1016/j.berh.2018.10.003
- Lakdawala N, Fedeles F. Vasculitis: Kids are not just little people. Clinics in dermatology. 2017;35(6):530-540. doi:10.1016/j.clindermatol.2017.08.004
- Lyons HS, Quick V, Sinclair AJ, Nagaraju S, Mollan SP. A new era for giant cell arteritis. Eye. 2020;34(6):1013-1026. doi:10.1038/s41433-019-0608-7
- Dasgupta B, Borg FA, Hassan N, et al. BSR and BHPR guidelines for the management of giant cell arteritis. Rheumatology. 2010;49(8):1594-1597. doi:10.1093/rheumatology/keq039a
- Chandran AK, Udayakumar PD, Crowson CS, Warrington KJ, Matteson EL. The incidence of giant cell arteritis in Olmsted County, Minnesota, over a 60-year period 1950-2009. Scandinavian journal of rheumatology. 2015;44(3):215-218. doi:10.3109/03009742.2014.982701
- Smith CA, Fidler WJ, Pinals RS. The epidemiology of giant cell arteritis. Report of a ten-year study in Shelby County, Tennessee. Arthritis and rheumatism. 1983;26(10):1214-1219. doi:10.1002/art.1780261007
- Aiello PD, Trautmann JC, McPhee TJ, Kunselman AR, Hunder GG. Visual prognosis in giant cell arteritis. Ophthalmology. 1993;100(4):550-555. doi:10.1016/s0161-6420(93)31608-8
- Salvarani C, Cimino L, Macchioni P, et al. Risk factors for visual loss in an Italian population-based cohort of patients with giant cell arteritis. Arthritis and rheumatism. 2005;53(2):293-297. doi:10.1002/art.21075
- Luqmani R, Lee E, Singh S, et al. The Role of Ultrasound Compared to Biopsy of Temporal Arteries in the Diagnosis and Treatment of Giant Cell Arteritis (TABUL): a diagnostic accuracy and cost-effectiveness study. Health technology assessment (Winchester, England). 2016;20(90):1-238. doi:10.3310/hta20900
- Schmidt WA. Ultrasound in the diagnosis and management of giant cell arteritis. Rheumatology. 2018;57(suppl_2):ii22-ii31. doi:10.1093/rheumatology/kex461
- Karakostas P, Dejaco C, Behning C, Recker F, Schäfer VS. Point-of-care ultrasound enables diagnosis of giant cell arteritis with a modern innovative handheld probe. Rheumatology. 2021;60(9):4434-4436. doi:10.1093/rheumatology/keab424
- Roncato, F.C., Gautier, G., Ploton, G., Denis, G., Espitia, O. and Roncato, C., 2020. Colour Doppler ultrasound of temporal arteries for the diagnosis of giant cell arteritis: a multicentre deep learning study. Clinical and experimental rheumatology.
- Qasrawi, R., Al-Halawa, D.A., Daraghmeh, O., Hjouj, M. and Seir, R.A., 2021. Medical Image Processing and Analysis Techniques for Detecting Giant Cell Arteritis. In Giant-Cell Arteritis. IntechOpen.
- McMaster C, Yang V, Sutu B, et al. Temporal artery biopsy reports can be accurately classified by artificial intelligence [abstract]. Arthritis Rheumatol 2021;73 Suppl. URL: https://acrabstracts.org/abstract/temporal-artery-biopsy-reports-can-be-accurately-classified-by-artificial-intelligence/
- Andersen, J.K.H., Pedersen, J.S., Laursen, M.S., Holtz, K., Grauslund, J., Savarimuthu, T.R. and Just, S.A., 2019. Neural networks for automatic scoring of arthritis disease activity on ultrasound images. RMD open, 5(1), p.e000891.
- Bressem, K.K., Vahldiek, J.L., Adams, L., Niehues, S.M., Haibel, H., Rodriguez, V.R., Torgutalp, M., Protopopov, M., Proft, F., Rademacher, J. and Sieper, J., 2021. Deep learning for detection of radiographic sacroiliitis: achieving expert-level performance. Arthritis Research & Therapy, 23, pp.1-10.
- Gal, Y. and Ghahramani, Z., 2016, June. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. In international conference on machine learning (pp. 1050-1059). PMLR.
- Ancona, M., Ceolini, E., Öztireli, C. and Gross, M., 2018, February. Towards better understanding of gradient-based attribution methods for Deep Neural Networks. In International Conference on Learning Representations (ICLR).
- Alber, M., Lapuschkin, S., Seegerer, P., Hägele, M., Schütt, K.T., Montavon, G., Samek, W., Müller, K.R., Dähne, S. and Kindermans, P.J., 2019. iNNvestigate neural networks!. J. Mach. Learn. Res., 20(93), pp.1-8.
Interested, please contact Prof. Dr. Shadi Albarqouni