MA Thesis: Deep Learning for Brain MRI Image Quality Transfer -- Not available
 © Shadi Albarqouni
© Shadi Albarqouni
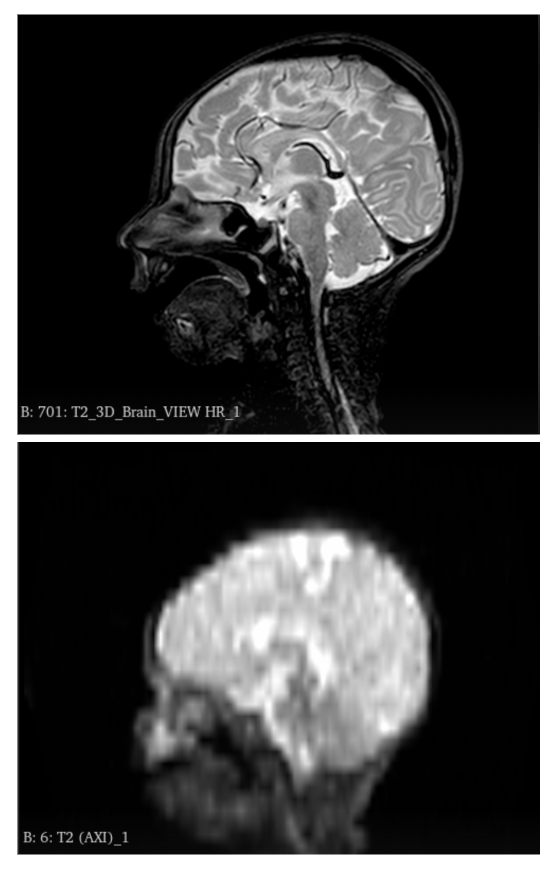
Abstract. Magnetic Resonance Imaging (MRI) plays a vital role in modern diagnostics, offering detailed, non-invasive insights into human anatomy [1-2]. High-field MRI (HF-MRI) systems, which operate at higher magnetic field strengths, provide superior image resolution and contrast compared to low-field MRI (LF-MRI) systems [3-4]. However, HF-MRI scanners are expensive and require significant maintenance, limiting their availability in resource-constrained environments where LF-MRI systems prevail. This disparity often leads to suboptimal diagnostic outcomes, underscoring the need to enhance the quality of LF-MRI images to the standard of HF-MRI [10].
Generative models, particularly diffusion models [5], have emerged as promising tools for image enhancement. They excel in generating high-fidelity, contextually coherent images by iteratively refining noisy data. Recent studies have demonstrated the potential of diffusion models in various imaging tasks, outperforming traditional techniques, including Generative Adversarial Networks (GANs). The proposed project aims to leverage diffusion models to improve LF-MRI image quality, bridging the gap between LF-MRI and HF-MRI and enhancing diagnostic accuracy [1-2].
The project aims to develop a 3D Conditional Diffusion Model specifically designed for enhancing LF-MRI images [6-8]. This model, named MagIQT, will integrate multiple innovative components that complement each other to address existing challenges in MRI image enhancement. The model will consist of a transformer-based encoder and a cross-batch mechanism, uniquely positioned to improve contextual information and reduce artifacts, thus advancing the current state-of-the-art. A recent review [9] summarizing the strengths and weaknesses of diffusion models in medical imaging will be further investigated.
Research Questions:
-
Q1) How effectively can a 3D conditional diffusion model enhance low-field MRI images to high-field MRI quality in terms of diagnostic accuracy and image fidelity compared to existing generative models [11]?
-
Q2) What are the specific contributions of the transformer-based encoder and cross-batch mechanism to the overall performance of the 3D conditional diffusion model in reducing artifacts and preserving anatomical details in MRI image enhancement?
-
Q3) How does the application of the 3D conditional diffusion model impact the detection and diagnosis of anomalies in pediatric MRI scans, particularly in identifying subtle pathologies that may be missed in low-field MRI?
Dataset: A total of 50 patients were scanned on portable 64mT and standard 3T scanners at the Clinic for Neonatology and Pediatric Intensive Care Medicine at the University Hospital Bonn with T1-weighted, T2-weighted, and FLAIR acquisitions. Brain Imaging sequences require registration as a pre-processing step. Another cohort of unpaired 100 patients was scanned on either 64mT or 3T, which might be used for the model development. Additional datasets might be requested from the authors of [11-12].
Roadmap:
- Familiarize yourself with the current literature [5-9]
- Build the baseline supervised model and develop the anomaly detection model.
- Run the necessary comparisons.
- Run extensive experiments and analysis
- Write up your thesis
Requirements:
- Solid background in Machine/Deep Learning
- Familiar with deep learning models and SOTA architectures
- Sufficient knowledge of Python programming language and libraries (Scikit-learn)
- Experience with a mainstream deep learning framework such as PyTorch.
- Machine/Deep learning hands-on experience
References:
- Barkovich, A. James. Pediatric neuroimaging. Lippincott Williams & Wilkins, 2005.
- Atlas, Scott W., ed. Magnetic resonance imaging of the brain and spine. Vol. 1. Lippincott Williams & Wilkins, 2009.
- Campbell‐Washburn, Adrienne E., et al. “Low‐field MRI: a report on the 2022 ISMRM workshop.” Magnetic resonance in medicine 90.4 (2023): 1682-1694.
- Arnold, Thomas Campbell, et al. “Low‐field MRI: clinical promise and challenges.” Journal of Magnetic Resonance Imaging 57.1 (2023): 25-44.
- Ho, Jonathan, Ajay Jain, and Pieter Abbeel. “Denoising diffusion probabilistic models.” Advances in neural information processing systems 33 (2020): 6840-6851.
- Kim, S., et al. “A 3D Conditional Diffusion Model for Image Quality Transfer - An Application to Low-Field MRI.” 1st Workshop on Deep Generative Models for Health at NeurIPS 2023. Link .
- Cechnicka, S., et al. “Ultra-Resolution Cascaded Diffusion Model for Gigapixel Image Synthesis in Histopathology.” NeurIPS 2023. Link
- Islam, Kh Tohidul, et al. “Improving portable low-field MRI image quality through image-to-image translation using paired low-and high-field images.” Scientific Reports 13.1 (2023): 21183. [Link] ( https://github.com/khtohidulislam/LoHiResGAN).
- Kazerouni, Amirhossein, et al. “Diffusion models in medical imaging: A comprehensive survey.” Medical Image Analysis 88 (2023): 102846.
- Kimberly, W. Taylor, et al. “Brain imaging with portable low-field MRI.” Nature reviews bioengineering 1.9 (2023): 617-630.
- Lucas, Alfredo, et al. “Multi-contrast high-field quality image synthesis for portable low-field MRI using generative adversarial networks and paired data.” medRxiv (2023).
- Arnold, T. Campbell, et al. “Sensitivity of portable low-field magnetic resonance imaging for multiple sclerosis lesions.” NeuroImage: Clinical 35 (2022): 103101.
Interested, please contact Prof. Dr. Shadi Albarqouni